or……
Quantitative microbiologcal analysis of cariogenic bacteria

In this part of the project, we wanted to find out more about all the things that “grow“ “live” in our mouths.
According to our sources, more than 300 different types of bacteria can be found in the oral cavity – the first contamination taking place in the birth canal. After the first teeth break through our gums, the acidogenic streptococci (of the species Streptococcus mutans), which are also responsible for caries, can be observed. This type of bacteria is not naturally part of the oral microflora. It is passed on through the parents’ saliva.
We focused on four major bacterial strains which are known to be the main cause for inflammatory tooth problems and gingivitis:
|
Streptococcus mutans
Streptococcus mutans is part of the streptococcal family. It is the major cause of tooth decay and can be detected in the mouths of many people. The level of concentration of Streptococcus mutans in the saliva correlates closely with the risk of caries. Streptococcus mutans is gram-positive and facultatively anaerobe. When growing on a culture medium, Streptococcus mutans forms roundish, convex colonies with a grained surface. This bacterium forms a biofilm, also known as plaque, which adheres to the teeth’s surface and feeds on various carbohydrates. While metabolizing sugar and other nutrients, it releases acids that cause tooth cavities. |
|
Our results were quite surprising!
|
Fusobacterium nucleatum (periodonticum, alocis, simiae)
Fusobacterium nucleatum is an anaerobe, gram-negative bacillus which can be found as a frequent but natural part of the oral microflora of humans.
An exceeding concentration of this bacterium may cause infections in the cervix ??? (gebärmutterschleimhaut?), the respiratory tract, and even in the liver.
Porphyromonas gingivalis
Porphyromonas gingivalis is gram-negative, anaerobe, and usually found in patients afflicted with gingival lesions. It is difficult to attest its presence in the saliva from a healthy mouth. According to literature, Porphyromonas gingivalis makes up 5% of the total microflora in an oral cavity with gingival lesions or advanced parodontitis.
Micromonas (Peptostreptococcus) micros
Micromonas micros is gram-positive, aero-tolerantly anaerobe, and directly linked with parodontitis. This bacterium is an early indicator for inflammatory gingivitis.
We intended not only to prove the presence of these different bacteria, but, furthermore, we wanted to show their level of concentration in the mouths of every participating pupil using RT-PCR (Real-Time Polymerase Chain Reaction).
Real-Time PCR is based on proving and quantifying the PCR-products during the amplification (multiplication of the starting material). Special dyes, whose fluorescence is directly proportional to the amount of PCR-products, help to make clear, quantitative statements concerning the amount of the starting material.
 |
The first step of this procedure was to isolate the genetic material, the DNA, of the bacteria.
We collected 5ml of saliva from each student in a sterile test tube.
500µl of each sample are transferred into a sterile 1,5ml reaction tube and the bacteria are concentrated by centrifugation.
The remaining 450µl of supernatant were disposed
The solid residue containing the bacteria (Pellet) is homogeneously mixed with 500µl lysis buffer. This buffer causes many cells to break and therefore the DNA is released. In order to efficiently break the cell walls of the gram-negative bacteria, we added 1mg/ml of lysozyme. To destroy the walls of the gram-positive bacteria, we treated the solution with 1 mg/ml mutanolysine.
|
| Primer, SYBRE Green® and flurophore for the mastermix |
We incubated the reaction for 30 minutes at 30°C, then for another 30 minutes at 60°C in the heating block. This incubation breaks the cell walls all the microorganisms present.
The extraction of bacterial DNA is carried out after vortexing and boiling the mixture at 100°C for 10 minutes. The sample is kept on ice.
In order to quantify the bacterial DNA, we chose a certain sequence of the double-stranded genome to be amplified via PCR.
The procedure was as follows:
Firstly, the DNA double-helix is denatured by boiling at 95°C. The two single strands are separated.
Then the primers (short strands of DNA which enable the polymerase, an enzyme able to duplicate the DNA, to bind to the DNA strands) attach themselves to the single strands of DNA, called annealing
Thirdly the double-helix is reconstructed by the polymerase, called extension
This cycle is repeated 30-40 times, leading to a multiplication of the DNA by the factor of x30 to x40.
In order to detect each pathogenic organism contained in the saliva samples, we had to prepare a suitable “mastermix”. This solution consisted of the following components:
• 15 µl iQ SYBR Green Supermix
(produced by BioRad)
• 1 µl primer 1
• 1 µl primer 2
• 11 µl sterile water |
|
DNA, TaqMan and polymerase for
the RT-PCR
|
Then we added 2 µl of the template, i.e. the total volume of the samples was 30 µl.
The primers were selected according to the DNA sequences of the respective bacterium we wanted to detect. ”SYBR Green“ places itself between the emerging DNA double-strands. After intercalation into a DNA double strand the dye emits light, making a real-time observation of the formation of new material possible during the PCR reaction.
The PCR apparatus we used (iCycler by BioRad), is able to analyze 96 samples at a time. The DNA samples of each pupil, as well as positive and negative control samples, were analysed in microtitre plates.
RT-PCR differs from conventional PCR inasmuch that it includes a quantitative analysis.
| The quantitative increase of the DNA is displayed real-time on the computer. This increase is measured via the emission spectrum of the dyes in the samples. In short, the more the sample glows, the more DNA it contains. This increase in light emission is visualized as a graph, which starts to rise from a certain PCR cycle onwards. This point is called Ct-value (critical threshold). Samples with a low Ct-value contain a high concentration of the micro organisms’ DNA and those with a high Ct-value contain only little DNA. The correlation between the Ct-value and the amount of initial material is quite relevant. A difference in the Ct-value of 1 reflects twice the amount of starting material; a difference of 2 means that the amount of starting material was 4 times higher and a difference of 5 indicated an initial concentration x32. |
|
Fig. 1
The results of 96 samples
|
Our first result was difficult to interpret as the negative control samples, used instead of templates (the DNA, we had isolated in the first place) contained only water, but yielded a positive signal. This indicated some form of contamination of our control samples, leading to a so called false-positive result.
Due to these results we prepared all PCR reactions in the sterile hood. Additionally, we sterilized the distilled water for the blank sample and the mastermix using UV light – which destroyed any DNA in the water.
 |
|
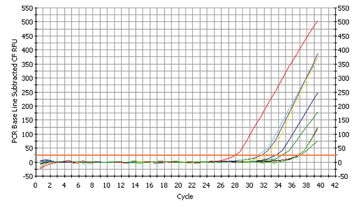
Fig. 2
The proof for Streptococcus mutans
|
Fig 2b
The initials of the pupils are given in the legend
|
This time the extra precautions paid off: The negative control samples remained negative, i.e. we could not detect the presence of a new product - we could trust the results this time and started with the analysis.
As can easily imagined by looking at the graph, the analysis of the data turned out to be quite difficult as the data output of the apparatus was enormous: (cp. fig. 1)
We tried to separate the single graphs by pupil and bacteria and made an exciting discovery:
16 pupils participated in the analysis.
The most dangerous cariogenic bacterium, Streptococcus mutans, was found in 7 of the 16 samples (cp. fig. 2a). The relative quantification of the converted Ct-values resulted in the following graph (fig. 2b):
 |
|
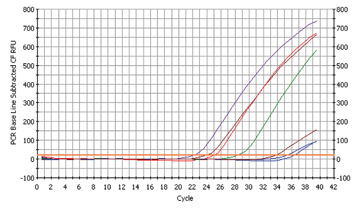
Fig. 3a
The proof for Fusobacterium nucleatum
|
Fig. 3b
The initials of the pupils are given in the legend
|
Only one pupil seemed to have a very high concentration of streptococci in the mouth with a Ct-value of 3,3 which is equivalent to a concentration x10 (cp. BK with PL). If the samples of pupil BK are compared with that of pupil SE, this makes a 256-fold higher concentration of this strain on the teeth! The fact that only few pupils were affected led us to the following, reassuring conclusion: conventional dentifrices are suited to impede the multiplication of the cariogenic bacterium Streptococcus mutans.
We found six samples to be contaminated with Fusobacterium nucleatum (cp. fig. 3a)
The Ct-values of Fusobacterium nulceatum were far lower than those of Streptococcus mutans, indicating that this bacterium is more prevalent in the oral cavity.
 |
|
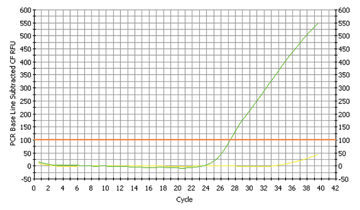
Fig. 4a
The proof for Porphyromonas gingivalis |
Fig. 4b
The initials of the pupils are given in the legend |
To speak in figures: The sample with the highest concentration had 60 times more bacteria than the sample with the highest concentration of Streptococcus mutans! (cp. fig. 3b)
When looking closer at this bacterium we realized that it was very common in connection with sore throats. The pupil with the highest concentration of Fusobacterium had been suffering from tonsilitis shortly before our day in the laboratory! This was a fascinating proof for the reliability of our method!
 |
|
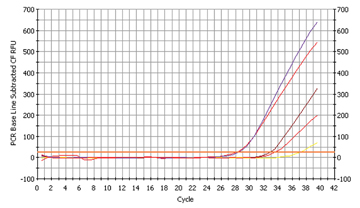
Fig. 5a
The proof for Micromonas micros
|
Fig. 5b
The initials of the pupils are given in the legend
|
Only one sample contained a high concentration of Porphyromonas gingivalis (cp. fig. 4a and fig. 4b). The concentration of Porphyromonas was in between the two other samples of Fusobacterium and Streptococcus, i.e. 8 times smaller than Fusobacterium, but 8 times larger than Streptococcus. According to our literature sources, this bacterium is very rare in a healthy oral microflora, but can multiply enormously, if the human being is suffering from gingivitis. And again our results were surprising: The student with the positive sample had just cut one of his wisdom teeth and was suffering from gingivitis!
The fourth bacterium we wanted to detect, Micromonas micros, has not been subject to extensive studies compared to the other three bacteria. It is known to occur frequently in the earlier stages of parodontitis. We found out that two of our students run the risk of suffering from gingival diseases (fig. 5a and fig. 5b) – a discovery which they ought to talk over with their dentists!.
|
